Archive detail
Unique identifiers for mass spectra
November 24, 2016 |
Mass spectrometry is a highly sensitive analytical technique which makes it possible to detect minute quantities of substances – even within mixtures. The technique is so powerful that it can detect the equivalent of a sugar lump dissolved in a swimming pool. As well as identifying known compounds, mass spectrometry is also used to elucidate the structure of newly discovered compounds. Since the development of the first commercial mass spectrometer in the 1950s, systems and methods have been continuously optimized, and mass spectrometry has now become an indispensable analytical tool for basic chemical and biological research, environmental and climate science, medicine, and forensics.
Taming the profusion of data
Every day, gigabytes of mass spectral data are collected by scientists around the world. Millions of spectra – amounting to several million gigabytes of data – are currently stored in around 20 major databases. Among these are thousands of reference spectra of known compounds, which are used for purposes of comparison. However, the databases also include mass spectra of as yet unknown compounds, now increasingly obtained from plants, fungi and marine organisms. In each case, these are stored in a database-specific format; consequently, for an unknown and unnamed substance X, it cannot be readily determined whether the compound has already been described and stored as a mass spectrum elsewhere. This complicates the exchange of information among scientists, for example concerning important properties of the substance X. The adoption of SPLASH should help to tame the profusion of mass spectral data that has arisen over time.
The programs developed by the international SPLASH consortium can generate a hashed identifier for any existing mass spectrum. As a result, spectra are not only searchable online, but all the data available on the substance in question can be combined from different databases. For unknown compounds, the assignment of a spectral identifier provides an initial name, thus dramatically facilitating communication.
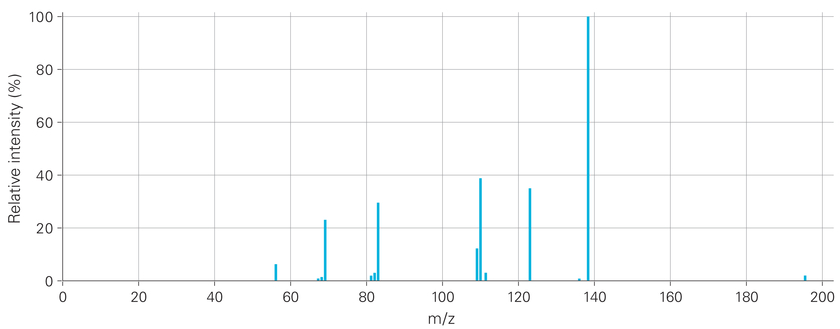
A typical mass spectrum of caffeine.
The full SPLASH reads: splash10‑000i‑3900000000‑73043667076aaf483c6e
http://mona.fiehnlab.ucdavis.edu/spectra/display/EA030313
Why is the development of identifiers so important?
Throughout the history of science, communication among chemists has been complicated by the fact that the same substances – depending where they were discovered or investigated – have been known by different names. Caffeine, for example, was initially named after the coffee plant (Coffea arabica) from which the compound was first isolated. It is also known as 1,3,7-trimethylxanthine, methyltheobromine or theine.
In the early 20th century, standards for chemical nomenclature, symbols and terminology were developed by the International Union of Pure and Applied Chemistry (IUPAC); these are still applied worldwide today. The official IUPAC name for caffeine is 1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione. This systematic nomenclature is particularly useful for the naming of unknown compounds, although additional (trivial) names generally become established in everyday use.
While the IUPAC name is universally applicable and is understood by chemists worldwide, it has the disadvantage that – particularly in the case of complex compounds – it is too long to permit visualization of the spatial arrangement of the atoms in the compound. For this reason, chemists prefer the graphical representation of spatial relationships known as the structural formula. However, structural formulas are not readily intelligible to computers. Therefore, in order to facilitate web searches, IUPAC initiated the development of two algorithms for converting information on the structure of chemical compounds into machine-readable sequences of characters – the International Chemical Identifier (InChI) and the hashed InChI (InChIKey). Both of these identifiers can be generated for all existing compounds using freely available software. The InChI and InChIKey are now also included in public chemistry databases and portals such as PubChem or ChemSpider, as well as Wikipedia. If all or part of the identifier for caffeine is entered in a search engine, all the relevant pages on this compound will be found, including the structural formula and other information of interest for scientists.
Since each compound has not only a unique structural formula but also a specific mass spectrum, the development of SPLASH is a logical extension of the InChI, given the growing volumes of spectral data stored in different formats.
The text is based on a press release by Sylvia Pieplow (Leibniz Institute of Plant Biochemistry, Halle).