News Detail
DNA fragments reveal the variety of species in rivers
November 6, 2018 |
Every living thing leaves behind tiny traces of its genetic material, for example in the form of dead skin cells or excrement. If one now takes water samples and decodes the environmental DNA (also known as eDNA) therein, one knows which species cavort in which waters. One thus discovers rare species that literally fall through the net during normal testing. To be sure, this concept of eDNA has long been known. But: “Until now one could determine from eDNA whether a species is present or not. But how this species is distributed in the whole ecosystem was unknown,” says Luca Carraro. He is now an Eawag researcher, after finishing his doctorate in environmental hydrology this summer at the EPFL.
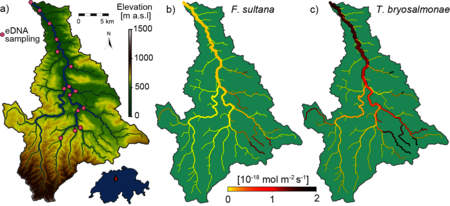
Together with researchers at Eawag, the ETH and the EPFL, he has succeeded in reconstructing the extensive presence of species from selective eDNA measurements. For the first time, Carraro and his colleagues have used the method to determine the biomass distribution of a bryozoan (Fredericella sultana) and its parasite (Tetracapsuloides bryosalmonae) in the Wigger – a prealpine Swiss river. The parasite causes the dreaded fish disease PKD, which carries off thousands of trout every year in Switzerland.
A colony of Fredericella sultana
(Photo: Michiel van der Waaij www.bryozoans.nl)
Computer model takes into account hydrology and ecology
In order to determine as exactly as possible the distribution of the organisms, the researchers developed a computer model based on hydrological and ecological concepts in river networks. Genetic material measurements are fed into the model and take into account the transport and decomposition of the eDNA fragments along the river, as well as local environmental factors. “This method will make it possible to monitor biodiversity all over the world simply and inexpensively. This was not possible up to now with conventional methods,” claims co-author and Eawag research group leader Hanna Hartikainen.
Location of the Wigger case study catchment area and the eDNA sampling points. b) and c) depict the model results of Fredericella sultana (b) and Tetracapsuloides bryosalmonae (c), reconstructed from eDNA measurements.
Original publication
Luca Carraro, Hanna Hartikainen, Jukka Jokela, Enrico Bertuzzo, and Andrea Rinaldo (2018). Estimating species distribution and abundance in river networks using environmental DNA. PNAS, https://doi.org/10.1073/pnas.1813843115