Department Fish Ecology and Evolution
Fish Genomics

Fascinated by the diversity of fish, this group aims to understand processes and factors creating variation and differentiation at the genomic level.
We analyse large-scale genomic data sets to gain a better understanding how processes like selection, drift, mutation, and recombination act in concert to create the observed genomic patterns of varying diversity and divergence along the genome. To achieve this aim we combine population genetics, bioinformatics, and an understanding of the study organisms (mainly fish).
Major themes
- Genome Evolution
- Speciation Genomics
- Ecological Genomics
Team
Selected publications
Current projects
Terminated projects
Stickleback Speciation Genomics

In this on-going large-scale genomics project we study the genome evolution at a population scale in an ecological and evolutionary model species, the three-spined stickleback. We analyse whole genome data from 66 three-spined sticklebacks, which originate from three ecotypes, a marine population and multiple lake-river population pairs, over a broad geographical range. Population diversity is contrasted between parapatric and geographically distant (allopatric) population pairs undergoing parallel ecological Adaptation.
Publications
Feulner, P.G.D.*, Chain, F.J.J.*, Panchal, M.*, Eizaguirre, C., Kalbe, M., Lenz, T.L., Mundry, M., Samonte-Padilla, I., Stoll, M., Milinski, M., Reusch, T.B.H., Bornberg-Bauer, E. (2013) Genome-wide patterns of standing genetic variation in a natural marine population of three-spined sticklebacks. Molecular Ecology 22: 635-49
Adaptive Introgression in Soay Sheep

Admixture, the mixing between divergent genomes, is widely thought to hinder local adaptation. However, a handful of recent studies have suggested that admixture can promote local adaptation. The Soay sheep of St Kilda are a primitive breed that have been the subject of a well documented long-term study, where data on life history, morphology, parasite burden, and pedigree information have been collected for over 7000 sheep. We followed up historical anecdotal evidence of an admixture event in the mid-late 1800s by screening 486 Soay sheep on a 50k ovine SNP chip. We found evidence for such a recent admixture event with a more modern, domesticated breed in the analysed genomic data. Utilising the HapMap dataset of over 60 different sheep breeds we showed that several haplotypes, previously demonstrated to be under selection in this population, have been introduced into Soay sheep from more modern breeds. Our study demonstrates that the introgression of domesticated alleles into wild populations is not necessarily disadvantageous and in fact it may provide a novel source of genetic variation capable of generating rapid evolutionary changes.
Publications
- Feulner, P.G.D. *, Gratten, J.*, Kijas, J.W., Visscher, P.M., Pemberton, J.M., Slate, J. (2013) Introgression and the fate of domesticated genes in a wild mammal population, Molecular Ecology 22: 4210-4221
Adaptive Radiation of African Mormyrids
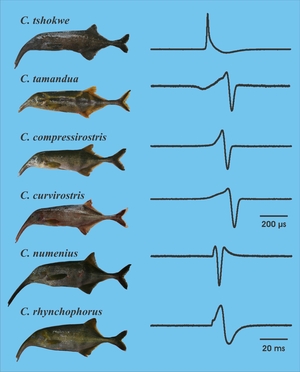
In this project we build a first comprehensive phylogeny of the mormyrid genus Campylomormyrus. We further assessed the importance of two very unusual features of these peculiar fish for the speciation process: their trunk-like elongated snout and their ability to produce electric signals for orientation and communication. Using morphometrics we demonstrated that the trunk morphology correlates with reproductively isolated groups characterised by specific electric signals, i.e. species. Behavioural studies further indicated that the waveform of the Electric Organ Discharge (EOD) causes assortative mating and therefore reproductive isolation. Because the EOD is also used for electrolocation of prey, we proposed the EOD as a ‘magic trait’ promoting the ecological speciation of Campylomormyrus within the Congo River.
Publications
- Feulner, P.G.D., Plath, M., Engelmann, J., Kirschbaum, F., Tiedemann, R. (2009) Electrifying love: electric fish use species-specific discharge for mate recognition. Biology Letters 5: 225-228.
- Feulner, P.G.D., Plath, M., Engelmann, J., Kirschbaum, F., Tiedemann, R. (2009) Article Addendum - Magic trait Electric Organ Discharge (EOD): Dual function of electric signals promotes speciation in African weakly electric fish. Communicative & Integrative Biology 2: issue 4.
- Feulner, P.G.D., Kirschbaum, F., Tiedemann, R. (2008) Adaptive radiation in the Congo River: An ecological speciation scenario for African weakly electric fish (Teleostei; Mormyridae; Campylomormyrus). Journal of Physiology – Paris 102: 340-346.
- Feulner, P.G.D., Kirschbaum, F., Mamonekene V., Ketmaier V., Tiedemann, R. (2007) Adaptive radiation in African weakly electric fish (Teleostei: Mormyridae: Campylomormyrus): a combined molecular and morphological approach. Journal of Evolutionary Biology 20: 403-414.
- Feulner, P.G.D., Kirschbaum, F., Schugardt C., Ketmaier V., Tiedemann, R. (2006) Electrophysiological and molecular genetic evidence for sympatrically occuring cryptic species in African weakly electric fishes (Teleostei: Mormyridae: Campylomormyrus). Molecular Phylogenetics and Evolution 39: 198-208.