Department Surface Waters - Research and Management
Antibiotic resistance as an emerging environmental contaminant
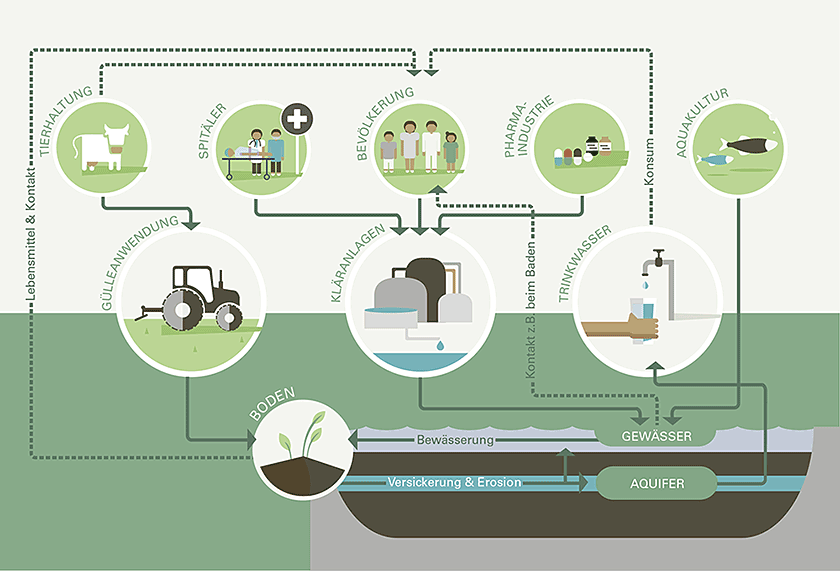
Antibiotic resistance is on the rise, and this represents one of the greatest biological risks of our time. Environmental bacteria are a natural source of resistance genes. However, the environment is also increasingly contaminated by resistant bacteria released with wastewater or from agricultural sources.
We study how resistant bacteria and their resistance genes spread in the aquatic environment. We investigate the dynamics of resistance during wastewater treatment and study strategies for an improved elimination of resistant bacteria.
Current Project
Dynamics of antibiotic resistance during oxidative treatment of wastewater
Wastewater treatments plants (WWTP) discharge antibiotic resistant bacteria with the treated wastewater. In this project we quantify the extent to which different WWTP in Switzerland eliminate and discharge resistant bacteria and how this affects the receiving waters.
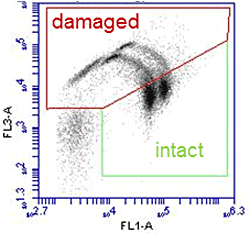
Oxidative wastewater treatment is an important strategy for the removal of micropollutants from wastewater, and is one of the main technologies that will be used for this purpose in Switzerland. In this Project we study how this treatment affects resistant bacteria. Laboratory studies investigate in detail how ozonation damages resistant cells and resistant genes to provide Information about dose dependence and kinetics of the process. Investigations at the first full scale ozonation plant in Switzerland, ARA Neugut, provide data from a real world implementation of this treatment process.
Collaboration
Urs von Gunten, Water Resources and Drinking Water, Eawag
Funding
BAfU project L361-1927
ResistFlow - Dynamics of the wastewater resistome during treatment
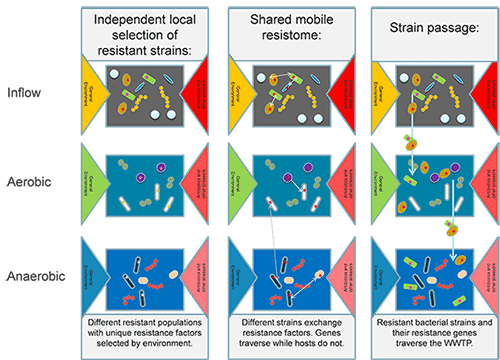
Anthropogenic release of antimicrobial resistant bacteria and resistance genes into environmental reservoirs, such as biological wastewater treatment plants (WWTPs), has come under scrutiny over its potential role in resistance dissemination. The mechanisms that allow resistance determinants to persist and reach the final effluent remain unknown. Learning about the in-situ expression of resistance genes in WWTP environments could provide important clues on the selective pressures that may determine their fate. This research project aims to address:
- how do resistance genes actually manage to traverse or even accumulate in WWTPs?
- are they actively expressed in response to selective pressure?
- do the micropollutants, including antibiotics and heavy metals, and operational conditions select for resistant bacteria and genes at each stage of secondary wastewater treatment?
In summary, this project provides paired metagenomic and metatranscriptomic points of views on the antimicrobial resistance in WWTPs. The outcomes will clarify the fate, expression level and co-selection of antimicrobial resistance, which promotes better understandings of the role and potential risk of wastewater treatment in the control and/or dissemination of resistance bacteria and genes, thus facilitating policy-making in the necessity for surveillance and/or control of these emerging anthropogenic contaminants after their release into downstream environmental reservoirs.
Contacts
Dr. Helmut Bürgmann, Dr. Feng Ju, Dr. Dave Johnson, Dr. Christa McArdell
Collaboration
Dr. Dave Johnson, Dr. Christa McArdell
Funding
Eawag Discretionary Funds
Concluded Projects
Microbial resistance, exotoxicological impact and risk assessment of micropollutants in a mid-sized lake
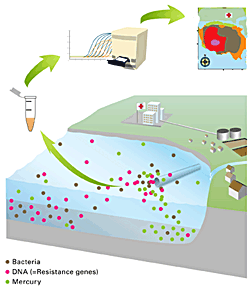
Antibiotics resistance is increasingly seen as an emerging environmental contaminant. In this project we studied antibiotic resistance in the wastewater stream of Lausanne, and the effect of the discharge of treated wastewater on the water and sediment of Lake Geneva, Switzerland.
Collaboration
Funding
Publications
- Bürgmann H. (2014) Eintrag von Antibiotika und Antibiotikaresistenzen in Wassersysteme der Schweiz. Prävention und Gesundheitsförderung 2014, 9:185–190, doi:10.1007/s11553-014-0444-3
- Czekalski N., Gascón Díez E., Bürgmann H. (2014) Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. The The ISME Journal (2014) 8, 1381–1390, doi:10.1038/ismej.2014.8
- Cantas L., Shah Syed Q.A., Cavaco L.M., Manaia C.M., Walsh F., Popowska M., Garelick H., Bürgmann H., Sørum H. (2013) A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Frontiers in Microbiology, May 2013, Vol. 4, Art. 96, 1-14, doi:10.3389/fmicb.2013.00096
- Czekalski N. (2013) Dissertation: Sources, Spreading and Fate of Antibiotic Resistance Genes and Resistant Bacteria in Vidy Bay, Lake Geneva, Switzerland, doi:10.5075/epfl-thesis-5637
- Czekalski N., Berthold T., Caucci S., Egli A., Bürgmann B. (2012) Increased levels of multiresistant bacteria and resistance genes after waste water treatment and their dissemination into Lake Geneva, Switzerland. Frontiers in Microbiology, March 2012, Vol. 3, Art. 106, doi:10.3389/fmicb.2012.00106
Antibiotic resistant microorganisms in the drinking water system of Lausanne

The occurrence of antibiotic resistant bacteria and resistance genes was studied in the drinking water production facilities and distribution network of Lausanne. Raw water, different treatment stages and the distribution network were analyzed over the course of one year.
Financing
Elimination of antibiotic resistant microorganisms from wastewater by membrane filtration

A pilot scale activated carbon powder – Ultrafiltration system was tested by the wastewater treatment plant of Lausanne. We investigated the prevalence of resistance at different stages of the WWTP and it’s temporal variability. The effectiveness of eliminating resistant bacteria by membrane filtration was determined.
Collaboration
Frederik Hammes, Environmental Microbiology, Eawag
