Department Environmental Microbiology
Microbial Specialized Metabolism
Research areas
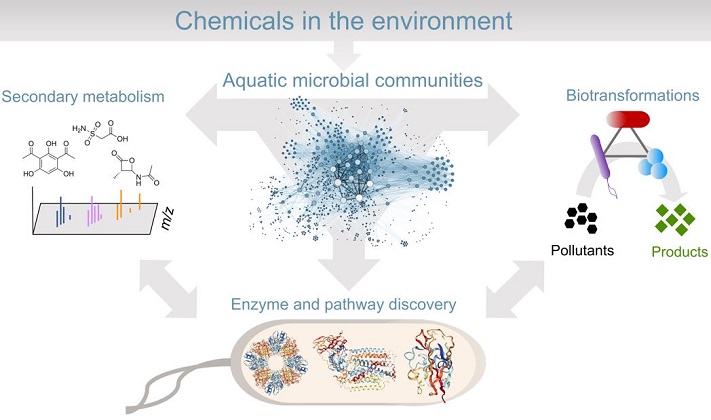
Microbes encode a remarkable diversity of enzymes to produce and break down chemical substances in the environment. In contrast to primary metabolism, we refer to 'specialized metabolism' here as pathways involving tightly-regulated enzymes often encoded by gene clusters for the synthesis, transformation, and degradation of bioactive compounds. Thanks to next-generation sequencing capabilities, we now see that we have only grazed the surface of the functional diversity of specialized enzymes and metabolites produced by uncultivated majority of microorganisms. In our group, we seek to answer the following research questions:
- What are the effects of anthropogenic inputs on the specialized metabolism of aquatic microbes in freshwater ecosystems?
- How do specialized metabolites affect biodegradation rates in microbial consortia?
- Can we use machine learning to predict the function and substrate specificity of specialized microbial enzymes involved in pollutant degradation?
For our current focus on the latter question, we use heterologous expression hosts and in vivo and in vitro assays to discover new enzymes and pathways from uncultivated microbes. We then use our experimental results to train machine learning models to learn patterns of substrate specificity and function of enzymes involved in secondary metabolite biosynthesis and pollutant biodegradation. In the long term, we are interested in characterizing and engineering enzymes as biocatalysts for environmental biotechnology applications such as improving water quality. At a more fundamental level, we are also interested in the ecological roles of specialized enzymes and metabolites in microbial communities.
For a full list of our publications, please visit Google Scholar
Selected publications
- Marti, T.D., Schärer M.R., Robinson, S.L. (2023) Microbial Biocatalysis within Us: The Underexplored Xenobiotic Biotranformation Potential of the Urinary Tract Microbiota. CHIMIA. doi: 10.2533/chimia.2023.424.
- Robinson, S.L.*, Piel, J., & Sunagawa, S. A. (2021) A roadmap for metagenomic enzyme discovery. Natural Product Reports. doi: 10.1039/D1NP00006C.
- Robinson, S.L., Biernath, T., Rosenthal, C., Young, D., Wackett, L.P., & Martinez-Vaz, B.M. (2021) Development of the organonitrogen biodegradation database: teaching bioinformatics and collaborative skills to undergraduates Journal of Biology & Microbiology Education. 22(1), ev22i1.2351. doi: 10.1128/jmbe.v22i1.2351.
- Wackett, L.P. & Robinson, S.L. (2020) The ever-expanding limits of enzyme catalysis and biodegradation: polyaromatic, polychlorinated, polyfluorinated, and polymeric compounds. Biochemical Journal. 477(15), 2875–2891. doi: 10.1042/BCJ20190720.
- Robinson, S.L., Smith, M.D., Richman, J.E., Aukema, K.G., & Wackett, L.P. (2020) Machine learning-based prediction of activity and substrate scope for OleA enzymes in the thiolase superfamily. Synthetic Biology. 5(1), ysaa004. doi: 10.1093/synbio/ysaa004.