News Detail
Spores archive antibiotic resistance
June 20, 2018 |
When living conditions for bacteria are unfavourable, some species make spores, which are able to survive for a long time under harsh conditions and be dispersed to other places. They are unharmed even at boiling temperatures for several hours. When environmental conditions improve, the spores awaken and become living bacteria once again, which divide and propagate. Whether such long-lived forms of bacteria can preserve and spread antibiotic resistance is the subject of research at Eawag and the University of Neuchâtel. The researchers have extracted bacteria spores from sediment drill cores in Lake Geneva and investigated their antibiotic-resistance genes. “We found a genuine archive of resistance genes in the sediment”, relates Helmut Bürgmann from Eawag.
Resistance genes in spores reflect antibiotic use
Because biomass and particles collect from year to year on the lake floor, one can date the different layers in a sediment drill core by year or by time period. The deeper one digs, the older the deposition. The researchers used two drill cores containing sediments from the last 100 years for their analyses. They extracted spores and bacterial cells from different layers of the drill core using a method developed by Assistant Professor Pilar Junier at the University of Neuchâtel. They then went on to determine the number of gene copies that code for resistance against two frequently used antibiotics – tetracycline and sulphonamide. As was expected, in the living bacteria cells the developmental history of the resistance genes was barely preserved, as they die in the sediment within a short time. In the spores, however, the resistance genes are still present years later. “The appearance of resistance spores shows a high correlation with antibiotic use over a period of time”, explains Bürgmann. For instance, the introduction of the antibiotic tetracycline in the 1960s was reflected in a rapid increase in resistance genes in bacteria spores in the corresponding sediment layers.
In addition to the time of appearance of resistance genes in bacteria spores, the method of spreading these genes was also of interest to the researchers. For this investigation they took nine sediment samples at different distances from the discharge point of the Vidy water treatment plant near Lausanne. As was the case in earlier experiments with bacteria cells, the researchers found the most resistance genes in the vicinity of the water-treatment plant. “Spores enter the lake by way of the treatment plant and sink into the sediment,” reports Bürgmann. Although the treatment plant is a good barrier for many intestinal bacteria, spores can possibly pass through more easily.
Intestinal bacteria as a reservoir
The researchers also investigated what type of bacteria the spores in the sediments belong to. They found that the frequency of resistance genes against tetracycline correlated strongly with the presence of the so-called Firmicutes bacteria, which are spore-generating bacteria found in the human digestive system. This result was observed in the Rhone delta as well as in the vicinity of the treatment-plant inlet in the Vidy bay. Especially dominant were representatives of clostridia, a subgroup of Firmicutes bacteria. “These results are an indication that the digestive system flora of humans is a reservoir for antibiotic resistance, which is spread widely by the spores”, says Pilar Junier. Spores of many different types of bacteria were on the contrary immune to sulphonamide. This confirms earlier observations that certain resistance genes can spread from one bacteria type to another. “Based on their longevity and the high dissemination rate, bacteria spores can contribute considerably to the spread and storage of antibiotic resistance in the environment”, concludes Bürgmann. He calculates that the probability of the spores in the lake sediments coming back to life is small, however.
Publications
Madueño, L., Paul, C., Junier, T., Bayrychenko, Z., Filippidou, S., Beck, K., … Junier, P. (2018). A historical legacy of antibiotic utilization on bacterial seed banks in sediments. PeerJ, 2018(1), e4197 (19 pp.). http://doi.org/10.7717/peerj.4197
Paul, Ch., Bayrychenko, Z., Junier, T., Filippidou, S., Beck, K., Bueche, M., Greub, G., Bürgmann, H., Junier, P. (2018). Dissemination of antibiotic resistance genes associated with the sporobiota in sediments impacted by wastewater. PeerJ 6:e4989; DOI 10.7717/peerj.4989
Publication excellence recognised
How can the spread of antibiotic resistance in the environment be reduced? Scientists from all over the world have combined their knowledge in a feature article – among them Helmut Bürgmann from Eawag. Their article “Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance“ was selected in March by the Environmental Science and Technology journal as the First Runner-up Feature 2017.
http://doi/10.1021/acs.est.7b03623
Strategies against the spread of resistant bacteria
Where do antibiotic-resistant bacteria eliminated in human faeces end up? There are many unresolved questions about whether and how such resistance is spread in various sanitation systems and through the reutilisation of faeces, urine and treated wastewater. There is also uncertainty as to how to evaluate the associated risks. Researchers in various institutes all over the world, including Eawag, have presented the current state of knowledge in a publication and highlighted where the gaps in our understanding lie. In addition, they outline how the battle against the proliferation of antibiotic resistance in sanitation systems and water can be fought globally. The researchers see improvements to sanitation and access to clean water as being particularly urgent.
Publication
Helmut Bürgmann, Dominic Frigon, William Gaze, Célia Manaia, Amy Pruden, Andrew C Singer, Barth Smets, Tong Zhang (2018). Water & Sanitation: An Essential Battlefront in the War on Antimicrobial Resistance. FEMS Microbiology Ecology, fiy101 https://doi.org/10.1093/femsec/fiy101
Images
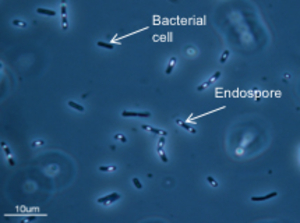
Bacteria with endospores (white grains). Certain bacteria can make spores and thus survive and disperse in inhospitable surroundings for a long time.
(Source: University of Neuchâtel)
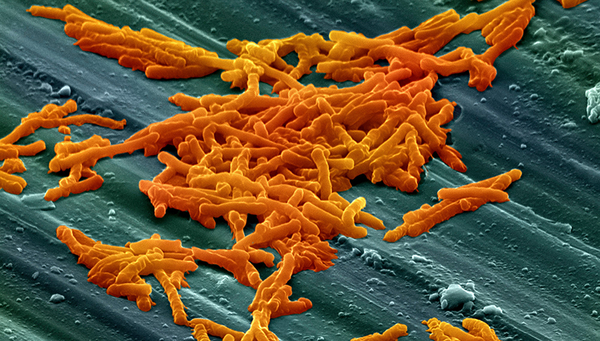
Bacteria spores with antibiotic-resistance genes are found in sediment drill cores in Lake Geneva. Their occurrence tallies with the use of antibiotics in the past.
(Source: University of Neuchâtel)
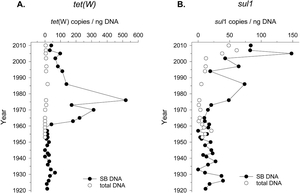
Relative frequency of antibiotic-resistance genes to tetracycline and sulphonamide in bacteria (total DNA) and bacteria spores in dated layers of the sediment drill core.
(Source: Madueño et al., 2018)
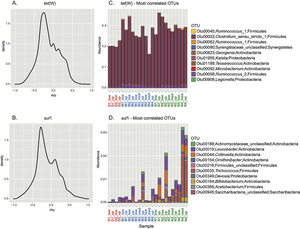
The frequency of spores from different bacteria types correlates with the occurrence of resistance genes: Tetracycline resistance can be assigned above all to Clostridium bacteria (above). Sulphonamide resistance correlates with different bacteria groups (below).
(Source: Paul et al., 2018)