Monitoring of SARS-CoV-2 in wastewater

Sampling at WWTP Werdhölzli, Zurich
(Photo: Esther Michel, Eawag)
The following answers to frequently asked questions on the detection of SARS-CoV-2 in municipal wastewater were jointly compiled by the departments of Urban Water Management and Environmental Microbiology at Eawag and the Environmental Chemistry Laboratory at EPFL.
FAQs
What is the significance of wastewater-based epidemiology in relation to SARS-CoV-2?
- Genetic material from the SARS-CoV-2 virus can be excreted in the faeces of infected individuals. This genetic material can be detected irrespective of whether the virus is still intact or only gene fragments are present.
- SARS-CoV-2 genetic material detected in wastewater provides evidence of infections in the population.
- The detection of SARS-CoV-2 in wastewater can help to define priorities for the regions concerned (e.g. with regard to materials, testing intensity, behavioural measures, etc.).
What contributions can wastewater-based epidemiology make?
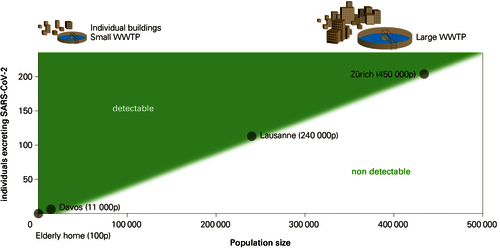
- Wastewater analyses allow large populations to be monitored (see Figure 1).
- A wastewater sample positive for SARS-CoV-2 can serve as an alert, triggering the adoption of measures in the region.
- Regular sampling can confirm – though not currently predict – the dynamics of an outbreak (increase in and levelling-off of infections). Data from the second wave in Zurich and Lausanne shows agreement between wastewater analyses and the case numbers reported on the basis of clinical tests (see the regularly updated graphs on the project website).
- Where large-scale clinical testing is no longer performed (e.g. after a significant decline in infections due to a vaccination campaign), wastewater analyses can provide evidence of residual or renewed circulation of the virus.
A wastewater sample positive for SARS-CoV-2 means that:
- recently recovered or currently infected individuals have contributed to the wastewater sample analysed.
- a threshold value of infected individuals has been exceeded (see Figure 1).
Figure 1. Conceptual model of the limit of detection for SARS-CoV-2 in wastewater as a function of the size of the population served by the wastewater treatment plant. For example, while it is possible to detect a very small number of individuals excreting SARS-CoV-2 in a home for the elderly with 100 people, the limit of detection at the Lausanne WWTP, which serves 240,000 people, is around 100 individuals excreting SARS-CoV-2 (in the case of Lausanne, based on the small number of clinical studies currently available, this would correspond to 10–20 new cases per day).
How is SARS-CoV-2 genetic material detected in wastewater?
- A sample of untreated sewage is generally collected from the inlet of a wastewater treatment plant (routine, flow-proportional 24h composite sample) and therefore encompasses a large number of toilet users in the catchment area of the wastewater treatment plant.
- The detection and quantification of SARS-CoV-2 in wastewater is based on real-time quantitative PCR (qPCR) or droplet digital PCR (ddPCR). These are the same RNA replication and analysis methods that are used for clinical tests.
- As the wastewater contains only very low concentrations of the target gene sequences (N1 and N2), the samples must undergo an elaborate preparation process involving filtration, concentration and RNA extraction prior to detection.
- For quality control, known concentrations of known viruses are also added to the samples. Differences between the amount of virus added and the amount of virus measured help inform us if and how well the measurements work.
How long does it take to detect SARS-CoV-2 genetic material in wastewater?
-
With the method currently used by EPFL and Eawag (for more information, see project website), the process of sample preparation, analysis and reporting takes two working days.
What can’t wastewater epidemiology do in relation to SARS-CoV-2?
- Wastewater samples do not reflect every positive case – due to the limits of detection (Figure 1) and because some infected individuals do not excrete SARS-CoV-2 genetic material.
- The analysis of wastewater samples does not distinguish between new cases of infection, on the one hand, and people who have recovered but are still excreting SARS-CoV-2 genetic material, on the other.
- Wastewater samples are no substitute for the swabs used to identify individual cases.
- Wastewater samples cannot be used to predict the further development of an outbreak.
- Wastewater samples can only provide indications of – but not confirm – whether the precautionary measures adopted have been successful (given the limits of detection, the impact of commuters and tourists or infected individuals who do not excrete SARS-CoV-2 genetic material).
Can mutations also be detected?
- If the RNA from wastewater samples is sequenced, mutations can also be found. This is, however, time-consuming and is not yet routinely performed. Thanks to sequencing, we were able – in collaboration with ETH and EPFL – to show that the British variant (B.1.1.7) was already present in Switzerland before the Christmas holidays. See the publication (preprint): Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples.
Are vaccines also detectable in wastewater?
- We are not aware of any studies that have sought to detect “residues” of COVID-19 vaccines in wastewater, and none of our projects are designed to do so. Experts believe it is unlikely that, for example, mRNA molecules from a vaccine are excreted in the faeces. And even if they did enter wastewater, they would not pose any risks, as they are not themselves biologically active and cannot self-replicate. Their function is merely to enable vaccinated persons to produce a protein, which is then recognised as part of the pathogen, triggering an immune response. In the body, the mRNA molecules are broken down within hours, and the proteins within days.
Why do the results vary so widely from day to day?
- Municipal wastewater is a complex mixture whose composition changes continuously, depending for example on weather conditions and the types of use in the catchment. The PCR reaction is often inhibited by organic compounds or metals in wastewater. Samples then have to be diluted, but only to such an extent that RNA detection is still possible. Over longer periods (several weeks), however, the SARS-CoV-2 curves from wastewater analysis are reliable and, above all, independent of people’s readiness to undergo testing or of any gaps in testing (e.g. during holidays).
Does the data from the six WWTPs indicate any regional peculiarities?
- Essentially, the SARS-CoV-2 curves from all the WWTPs reflect reported case numbers relatively well, and they show similar trends. Fortunately, the sharp rises observed between the beginning and middle of April have not continued.
- Periods showing systematic divergences can be found in the data from all six WWTPs. These need to be interpreted by experts from various disciplines – e.g. in the context of test positivity rates.
Will more WWTPs be added to the monitoring programme?
- We’ve been analysing samples from Zurich und Lausanne since the start of the second wave in October 2020 – first twice a week, then daily. Since February 2021, six WWTPs have been included in the FOPH project AbwasSARS-CoV-2: Zurich (Werdhölzli), Lausanne (Vidy), Laupen/BE (Sensetal), Altenrhein, Chur and Lugano. So altogether the programme covers almost a million residents, or around 11% of the Swiss population, in various regions.
- The project is to run until July 2021; we are currently discussing the question of a possible extension – and also expansion – with the federal authorities.
What does the project cost, and who is paying?
- The initial work was carried out as part of an SNSF project (Special Call: Coronaviruses); the analyses are currently being supported by the FOPH (approx. CHF 500,000) and the FOEN (approx. CHF 150,000), with substantial contributions from the research partners involved.
Certain cantons have started to analyse wastewater samples themselves. Are the results comparable?
- Eawag and EPFL warmly welcome the fact that cantons are using wastewater monitoring as one of a number of types of evidence for assessing the development of the pandemic. We are providing the relevant agencies with expert support, so as to ensure that the data series from various regions of Switzerland continue to be comparable.
- Publication of all data on a shared Web platform is currently under discussion. However, responsibility for coordination and implementation lies with the federal and cantonal authorities, not with researchers. One option would be to use, for this purpose, the Regional Laboratory Network, in accordance with Art. 18 of the Epidemics Act.
Can the virus spread via wastewater or even contaminate drinking water?
- According to the current state of scientific knowledge, the viruses in wastewater (i.e. in the WWTP inlet) are largely no longer active. The (fragile) viral envelope is damaged by various substances contained in wastewater (soap, etc.). To date, there have been no known cases of infection via untreated wastewater. In addition, initial results on elimination performance indicate that 99% of SARS-CoV-2 RNA fragments can be retained at WWTPs. This level is similar to that seen for many other viruses already present in wastewater before the emergence of the coronavirus. Nor is treated wastewater used directly as drinking water in Switzerland: it is first diluted in the environment and is only withdrawn after more or less prolonged residence times (with natural purification, e.g. by bank filtration); depending on local conditions, it may also undergo various treatment steps.
Are other countries also using wastewater analysis to track the pandemic?
- Yes. The number of research groups, authorities and individual cities using wastewater-based epidemiology as well as clinical testing to track the pandemic is constantly growing. An overview can be found at: https://www.covid19wbec.org/covidpoops19
What research questions are you currently focusing on?
- We are collaborating with ETH Zurich on using wastewater data to calculate the effective reproduction number (Re).
- In the longer term, the current project could provide a basis for developing wastewater-based epidemiology in Switzerland. This would make it possible to investigate other topics, such as the spread of influenza viruses or antibiotic resistance. That would be similar to the monitoring of drug use via wastewater samples, where the University of Lausanne and Eawag have contributed significantly to the development of pan-European standards.
Further information and links
Project website
Broadcast
SRF Einstein broadcast of 30 April 2020. “Der Lockdown: Das Virus und sein Impact” (Lockdown: the virus and its impact; 36 min).
News
- Projects by the Eawag Department of Urban Water Management in the area of source typing and tracking
- Eawag “Pathogens and Human Health” research group (Department of Environmental Microbiology)
- EPFL Environmental Chemistry Laboratory
Eawag/EPFL experts
Experts at Eawag and EPFL are Dr Christoph Ort, Dr Tim Julian and Prof. Dr Tamar Kohn.
Experts please send your enquiries by email to: abwasser.covid@cluttereawag.ch