Department Urban Water Management
Application of Wastewater-based Epidemiology for detection of respiratory viruses
The spread of infectious diseases is traditionally monitored with clinical data. However, for many pathogens, clinical data collection is limited. Wastewater-based surveillance offers a complementary approach to track infectious disease dynamics independent of clinical surveillance.
Weekly monitoring. Our aim is to inform infectious disease dynamics of circulating pathogens such as SARS-Cov-2, Influenza A & B and Respiratory Syncytial Virus (RSV) by surveilling influent wastewater from wastewater treatment plants throughout Switzerland. Since 2020, digital droplet PCR (ddPCR) assays have been developed and deployed to quantify viral RNA for multiple targets. The weekly produced data is published on the WISE dashboard (Wastewater Surveillance). We share findings from wastewater-based surveillance with the Federal Office of Public Health, which is responsible for infectious disease surveillance and responding to epidemics (see: IDD - Infectious Diseases Dashboard). Through our program, we help to support data-driven health policy decisions on infectious diseases in Switzerland.
Content and objectives of research projects. The aim of this project is to further develop the methods and analyses of wastewater-based infectious disease surveillance that we established during the Covid-19 pandemic. In particular, we extend our work to other pathogens (human coronaviruses, noroviruses and other enteric pathogens) and other methods (e.g. monitoring for antimicrobial resistant bacteria by plating and whole genome sequencing to detect signature mutations). In doing so, we seek to answer the following research questions:
- What factors determined the epidemiological spread of viruses? How early can virus outbreaks be detected and future spread modelled based on wastewater analyses?
- Can we quickly determine the genetic variants that start an epidemiological wave? Can we determine the future evolution of the wave based on wastewater data?
- How does a pandemic virus transform into an endemic virus? To what extent can we predict endemic characteristics?
History
Update January 2025: Showing that the reduction of the number of surveilled wastewater treatment plants (WWTPs) still captures the regional trends of viral load in wastewater without a significant information loss, we reduced the weekly monitored WWTP’s from 14 to 10. From now on, the WWTPs of Lugano, Zürich Werdhölzli, Lausanne Vidy, Chur, Sensetal Laupen, Basel, Geneva Aire, Luzern Emmen/Buholz, Altenrhein and Neuchatel get monitored on a weekly basis.
Update July 2023: In the context of the continuation of the national wastewater surveillance program, Eawag will perform all analyses and ramp up from six to 14 wastewater treatment plants. In the transition time between 29th June and 10th July a reduced number of samples or no samples will be processed due to adjusting logistics and performing additional quality assurance.
4. July 2023: In the context of the continuation of the national wastewater surveillance program, Eawag will perform all analyses and ramp up from six to 14 wastewater treatment plants. In the transition time between 29th June and 10th July a reduced number of samples or no samples will be processed due to adjusting logistics and performing additional quality assurance.
Starts in November 2022: Wastewater-based Infectious Disease Surveillance WISE SNSF Sinergia Project (in collaboration with ETHZ and EPFL)
The spread of infectious diseases is traditionally monitored with clinical data. However, for many pathogens, clinical data collection is insufficient. Wastewater-based epidemiology, which is the focus of this project, offers a complementary approach to track infectious disease dynamics independent of clinical surveillance.
Content and objectives of the research project. The aim of this project is to further develop the methods and analyses of wastewater-based infectious disease surveillance that we established during the Covid-19 pandemic. In particular, we will extend our work to other pathogens (human coronaviruses, influenza, noroviruses and other enteric pathogens). In doing so, we primarily want to answer the following three research questions:
- What factors determined the epidemiological spread of viruses? How early can virus outbreaks be detected and future spread modelled based on wastewater analyses?
- Can we quickly determine the genetic variants that start an epidemiological wave? Can we determine the future evolution of the wave based on wastewater data?
- How does a pandemic virus transform into an endemic virus? To what extent can we predict endemic characteristics?
Scientific and societal context of the research project. We will share findings from wastewater-based epidemiology directly with the Federal Office of Public Health, which is responsible for epidemics, and the relevant decision-makers. Thus, for the aspects monitored here, we provide an additional, data-driven foundation for health policy decisions. In general, we will support the transfer of research on wastewater-based epidemiology into a nationwide surveillance system.
Project descriptionWastewater based epidemiology provides insights into the disease burden and disease dynamics of the current COVID-19 pandemic. Through surveillance of wastewater for the presence of SARS-CoV-2 RNA at wastewater treatment plants throughout Switzerland, wastewater complements other epidemiological indicators, such as reported cases, to help inform COVID-19 disease dynamics. SARS-CoV-2 RNA in wastewater also provides insights into the circulation of Variants of Concern through targeted detection of signature mutations as well as through whole genome sequencing. In this project, we continue to develop, apply, and evaluate methods for sensitive and specific detection of SARS-CoV-2 and other pathogens in wastewater to help inform disease dynamics.
Project history
Update December 2021
We are monitoring the wastewater for the relative proportion (percentage) of signature mutations of the Delta Variant (S:L452R) and the Alpha/Omicron Variants (HV69/70). The data are available here and a visualization can be found here.
Update June 2021
Based on the wastewater data, our ETHZ project partners (Group cEvo, BSSE) estimated the effective reproductive number Re, i.e. the number of people an infected individual will infect over time. These estimates are visualized here with a direct comparison of estimates based on clinical cases.
Update May 2021
Extracts of wastewater samples are sequenced at the FGCZ. With bioinformatics tools, our ETHZ project partners (Group Computational Biology, BSSE) estimate the prevalence of different genomic variants of SARS-CoV-2. The results for six wastewater treatment plants for B.1.1.7, B.1.351, P.1 and B.1.617* are presented here.
Update April 2021
The AbwasSARS-CoV-2 Project launched in February 2021 in collaboration with the FOPH (see Point de Presse, 9 March 2021, Link zu Video from minute 15:36, German). In this project, we are collecting and quantifying SARS-CoV-2 RNA from six wastewater treatment plants across Switzerland daily (see Figure 2; results here).
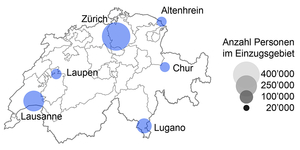
Figure 2: Locations of the six wastewater treatment plants being investigated in the FOPH's AbwasSARS-CoV-2 project, with information on the size of the catchment areas. Together, these treatment plants collect and treat the wastewater of almost 1 million inhabitants.
Updates are normally provided once or twice per week (depending on frequency of sample shipment and processing capacity not all locations are updated on the same day). The following links direct you to the data measured at the wastewater treatment plants (currently only in German):
Overview all wastewater treatment plants
Altenrhein ARA Altenrhein
Chur ARA Chur
Laupen ARA Sensetal
Lausanne STEP Vidy
Lugano CDA Lugano
Zürich ARA Werdhölzli
Update October 2020
After an intense period of method development and data collection, we are happy to now provide:
Near-real-time measurements for the two wastewater treatment plants, ARA Werdhölzli in Zürich and STEP Vidy in Lausanne: graph for Zurich and graph for Lausanne [these graphs are not updated any longer after the April update, see new links above]
Our protocols for sample preparation: ultrafiltration protocol (updated version here) and extraction protocol
Answers to frequently asked questions (see link to FAQ)
Original project description June 2020
Application of Wastewater-based Epidemiology to SARS-CoV-2 Detection
Wastewater provides potential insights into improved understanding of the disease burden and disease dynamics of the current COVID-19 pandemic. Although COVID-19 is a respiratory disease, a substantial portion of people shed signatures of the virus (RNA) in their feces. By collecting and analyzing wastewater samples for these signatures, we can determine whether or not, and to what extent, people within a wastewater treatment plant catchment area are infected with the virus. We may also be able to detect the virus circulating in communities via wastewater before clinical case data is available, due to the lag between symptoms and confirmed testing results.
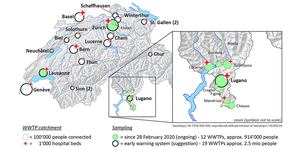
Figure 1. At the beginning of the first wave, we collected samples from 12 wastewater treatment plants (green circles) and continued with Lausanne and Zurich. Samples from 19 large wastewater treatment plants (bold circles) would cover 2.5 million people.
In this study, we (the laboratories of Tim Julian and Christoph Ort, Eawag; Tamar Kohn and Xavier Fernandez Cassi, EPFL) are analyzing wastewater influent samples from Lausanne, Zurich, and Ticino for the presence and quantity of SARS-CoV-2 from the start of the outbreak (late February) to the present. We will integrate the data into models of disease trajectory, in hopes that we can contribute to insights about the disease in Switzerland. Furthermore, we will develop and improve methods for the detection of SARS-CoV-2 at very low concentrations to help develop and improve an early warning system for the emergence or re-emergence of the disease.
Archived dashboards
Scientific Reports
Current reports can be found on the WISE project website under the heading ‘Reports’: Wastewater Surveillance
Videos & Print media
Current articles from print media and videos are available on the WISE project website under the heading «Printmedia» and «Videos»: Wastewater Surveillance