Department Environmental Microbiology
Microbial Specialized Metabolism
Research areas
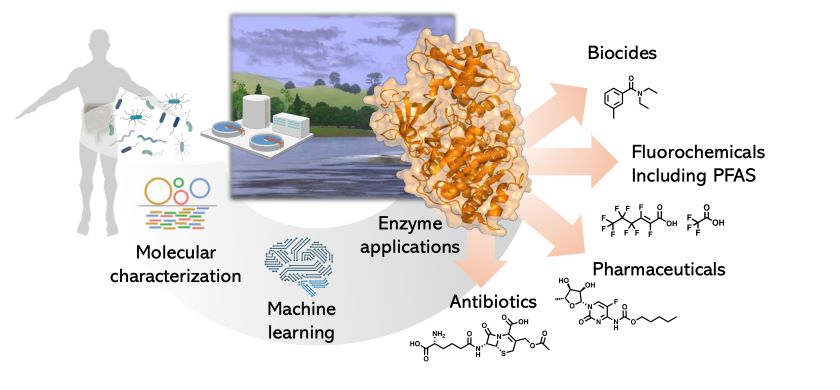
Our research group seeks to understand how naturally occurring microbes, including bacteria and fungi, can break down chemical pollutants. We use a combination of experiments and computational modeling to predict how different microbial enzymes biotransform industrial compounds, pharmaceuticals, and agrochemicals. A major focus in our group is on fluorinated compounds, specifically PFAS (Per- and Polyfluoroalkyl Substances), which are called 'forever chemicals' because of their persistence in the environment. Part of the reason for their resistance to degradation comes from the strong carbon-fluorine chemical bond. We've shown that some microbes, including bacteria from the human gut, have enzymes capable of breaking C-F bonds. However, these enzymes are only effective on simple fluorinated compounds with only one or two fluorine atoms, but not on long-chain, perfluorinated PFAS, which can contain fifteen or more fluorine atoms. We are working to improve this activity and learn how microbial enzymatic diversity can tackle the vast chemical diversity of fluorinated compounds. Ultimately, our long-term vision is to enhance our ability to understand, predict, and engineer microbial enzymes for pollutant detection and removal.
Another focus in our group is on microbial biosynthesis. We are interested in microbial bioproducts such as secondary metabolites and extracellular polymeric substances, as well as their ecological functions.
For more information, feel free to contact us directly!
For a full list of our publications, please visit Google Scholar